Case in point, here's a quote from a
recent paper at the
Journal of Human Genetics (emphasis is mine):
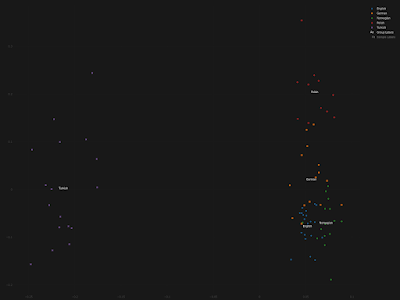
The Mordovian and Csango samples have a moderate to slight orientation toward the Central-Asian and Siberian Turkic groups. This could suggest the more significant East Eurasian or Turkic ancestry of these populations, which should be further investigated. German samples are inhomogeneous, and some of the German samples also show this tendency, which can be the result of the recent 20th century Turkish immigration into Germany [42].
Nope, these German samples don't show anything even remotely resembling recent Turkish ancestry. The authors of the paper, Ádám, V., Bánfai, Z., Maász, A. et al., should've been able to figure this out, even with the standard analyses that they ran on their dataset. Failing that, the peer reviewers at the
Journal of Human Genetics should've noticed that the authors were confused.
Moreover, if the authors and peer reviewers actually bothered to take a closer look at metadata for these samples, which were sourced from the
Estonian Biocentre, they'd see that they're not even from Germany. In fact, they represent self-reported ethnic Germans from Russia.
My own quick and dirty analysis of these individuals suggests that many of them harbor East Slavic and/or Volga Finnic ancestries. Indeed, only some of them can pass genetically for run of the mill Germans from Germany. The Principal Component Analysis (PCA) below is self-explanatory. It was run with the Vahaduo Custom PCA tools freely available
here. The relevant PCA datasheet can be gotten
here.
That's not to say, of course, that some Germans don't have recent Turkish ancestry, because an increasing number of Germans nowadays do, nor that people with German heritage in Russia shouldn't identify as Germans, because that's entirely their choice.
This blog post isn't about what it takes to be German, and this is not something that I ever want to discuss for obvious reasons. The point I'm making here is that the authors and peer reviewers of the said paper at the
Journal of Human Genetics were sloppy and half-arsed in their approach. And, sadly, this isn't an isolated case in peer reviewed scientific literature dealing with human population genetics.
I feel that the Estonian Biocentre is also partly to blame for this cock up, due to its somewhat peculiar sampling and labelling strategies. For instance, its scientists rely solely on self-reported identity to establish the ethnic origins of their samples, and they apparently never remove genetic outliers from their datasets or even try to identify them.
Unfortunately, I fear that this relaxed approach will eventually lead to basic errors and even unusual conclusions in a number of so called peer reviewed papers.
I first raised this issue with the Estonian Biocentre about five years ago, when I noticed that some of the supposedly Polish individuals in its dataset were genetically more similar to various groups from northern Russia than to Poles from Poland. These individuals also showed significant Siberian ancestry, which was very unusual indeed. Where the hell did the Estonian Biocentre find Poles who resembled people from near the Arctic circle, you might ask? Apparently in Estonia.
OK, I can imagine that sampling ethnic Poles from Estonia may have been easier for the Estonian Biocentre than sampling Poles from Poland. And Estonian Poles certainly make for interesting and useful data points. However, as you can see in the PCA below, some of these samples (labeled Polish_Estonia by me) aren't representative of the native Polish population, and yet the Estonian Biocentre not only lumps them with their Poles from Poland sample set, but even labels them with the word "Poland". The relevant PCA datasheet can be gotten
here.
But, based on my communications with some of the scientists at the Estonian Biocentre, including head honcho Mait Mestpalu, it seems that nothing will ever change there in regards to this issue. Who knows, perhaps some day we'll see a paper based on Estonian Biocentre data in the
Journal of Human Genetics claiming that Poles originated near the Arctic circle? I wouldn't be shocked if that actually happened.
Citation...
Ádám, V., Bánfai, Z., Maász, A. et al.
Investigating the genetic characteristics of the Csangos, a traditionally Hungarian speaking ethnic group residing in Romania. J Hum Genet (2020). https://doi.org/10.1038/s10038-020-0799-6
See also...
Like three peas in a pod